Performance of an atlas-based auto-segmentation software for MRI-only H&N cancer radiotherapy
PO-1668
Abstract
Performance of an atlas-based auto-segmentation software for MRI-only H&N cancer radiotherapy
Authors: Stefano Riga1, Chiara Carsana2, Marco Felisi1, Sibio Daniela2, Lizio Domenico1, Angelo Filippo Monti1, Roberto Giuseppe Pellegrini3, Denise Curto1, Muti Gaia1, Oscar Enrique Panchi Maigualca1, Barbara Bortolato2, Francesco Bracco2, Angelo Vanzulli4, Mauro Palazzi2, Alberto Torresin5
1ASST Grande Ospedale Metropolitano Niguarda, Medical Physics Department, Milan, Italy; 2ASST Grande Ospedale Metropolitano Niguarda, Radiotherapy Department, Milan, Italy; 3Elekta AB, Global Clinical Science, Stockholm, Sweden; 4ASST Grande Ospedale Metropolitano Niguarda, Radiology Department, Milan, Italy; 5ASST Grande Ospedale Metropolitano Niguarda,, Medical Physics Department, Milan, Italy
Show Affiliations
Hide Affiliations
Purpose or Objective
In recent years, there has been
an increasing interest in the application of magnetic resonance (MR) in
radiation therapy (RT), essentially due to the benefits that using magnetic
resonance imaging (MRI) provides. MRI allows an optimal soft tissue
differentiation of target tumours and organs-at-risk (OARs). Moreover, MRI-only
RT is desirable when repeated scanning can be helpful during treatment to
monitor early response and evaluate possible OARs and target changes, i.e., for
replanning during treatment while reducing patient X-ray exposures. Accurate
OARs delineation is of critical importance to achieve the most efficient radiation
planning process, mainly to ensure adequate dose coverage whilst minimising
dose to OARs. However, manual delineation of each structure is a time-consuming
effort. Moreover, this segmentation is subject to observer bias and
intra/interobserver variability. While that is still the most widely used
approach, software-based auto-segmentation techniques are being increasingly
used clinically. This work aims to evaluate the use of ADMIRE® software
(research version 3.28, Elekta AB, Sweden) for the multi-atlas-based
segmentation of H&N structures on MR images.
Material and Methods
To create a multi-atlas database,
the MRI scans of eleven subjects were acquired using a T1w 3D VIBE Dixon
gradient echo sequence on a 1.5T Magnetom Aera scanner (Siemens Healthcare, Germany).
The MRI sequences were acquired in the same setup of CT simulation, including a
thermoplastic mask fixed on a flat table. Twenty-six structures (OARs, bone and
air) were contoured on each MRI dataset with the support of a radiation
oncologist and a neuroradiologist. The geometric accuracy of three different algorithms
implemented on ADMIRE® (STAPLE, Patch Fusion and Random Forest) was evaluated
in terms of Dice similarity coefficient (DSC), using a leave-one-out
cross-validation approach and considering the OAR manually segmented as the
gold standard.
Results
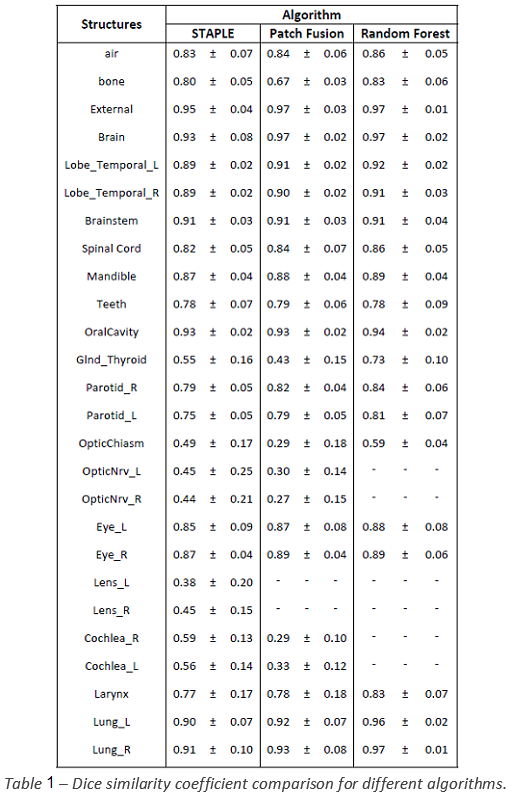
The mean DSC (± 1 std. dev.) of
the different methods were reported in Tab. 1. The Random Forest (RF) algorithm
showed better results compared to the other two and proved to be a reliable
tool for automatic delineation. Nevertheless, the RF algorithm has not proven
capable of correctly contouring small structures (lens, cochlea and optical
nerves). However, the time required for contouring these structures is significantly
short and has almost no impact on the total contouring time. An example
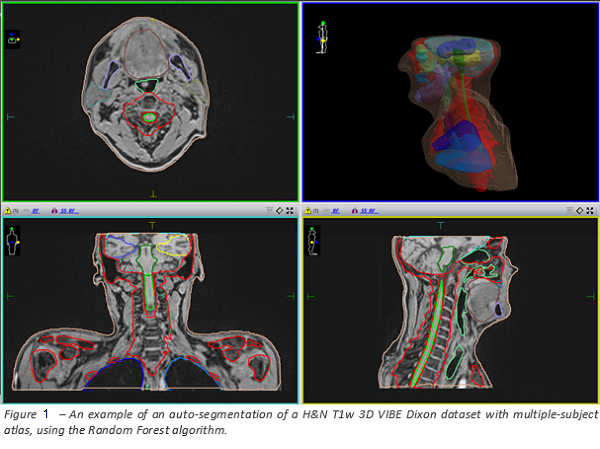
auto-contouring result of the Random Forest algorithm is reported in Figure 1.
Conclusion
This
study demonstrated the feasibility of accurate atlas-based automatic
OARs segmentation that could lead to significant time-saving in the overall
MRI-only RT workflow. Therefore, improvements in the automatic segmentation of
these OARs are required to reduce the time of the manual editing process, and
further investigations are necessary to improve its performance for small
structures.