Automatic segmentation of individual lymph nodes in head and neck cancer patients using 3D CNNs
PO-1593
Abstract
Automatic segmentation of individual lymph nodes in head and neck cancer patients using 3D CNNs
Authors: Floris Reinders1, Mark Savanije1, Chris Terhaard1, Patricia Doornaert1, Cornelis van den Berg1, Cornelis Raaijmakers1, Marielle Philippens1
1University Medical Centre Utrecht, Radiotherapy, Utrecht, The Netherlands
Show Affiliations
Hide Affiliations
Purpose or Objective
Irradiation
of individual lymph nodes (i-LNs) instead of conventional lymph node levels in head
and neck cancer (HNC) patients reduces the radiation dose to nearby organs at
risk, potentially leading to less radiation induced toxicity. Since contouring
of all i-LNs is very time-consuming, 2 convolutional neural networks (CNNs)
were trained, tested and compared for the automatic segmentation of i-LNs and
LN levels on MRI.
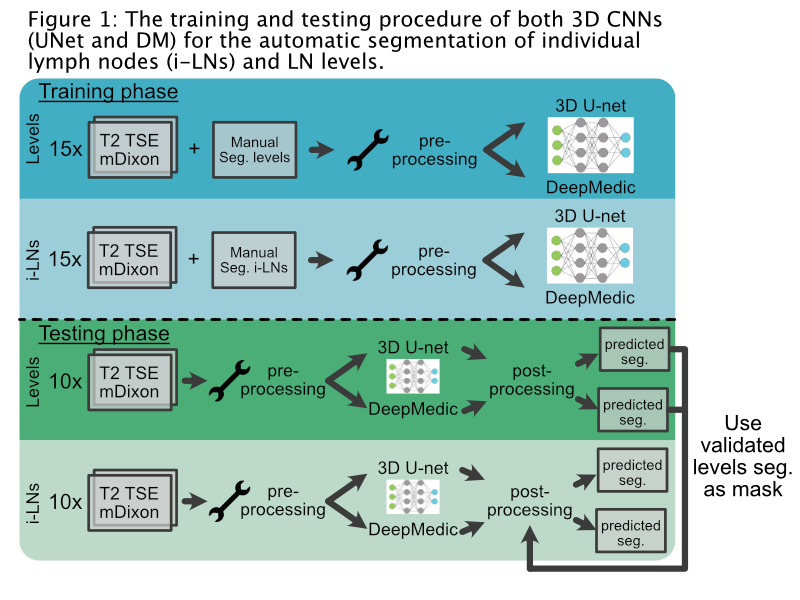
Material and Methods
Multiple
Dixon T2-weighted turbo spin echo (T2 mDixon TSE) MRI scans of 25 head and neck
cancer patients were used for manual contouring of i-LNs and LN levels
(Ib-II-III-IVa-V) as reference. The water image and the in-phase image of the
T2 mDixon TSE were used as input channels.
Pre-processing
was done by normalization, clipping at the 99th percentile and
resampling to 1 mm³ of all images. Two 3D
convolutional neural networks (nnU-net (UNet) and DeepMedic (DM)) were trained with the scans
of 15 patients. During post-processing the automatically segmented LN levels
were, after manual confirmation, used as a mask to select only i-LNs segmented
inside the LN levels. The MRI scans of 10 other patients were used for testing
both networks (Fig. 1) with manual contours as reference.
Testing
metrics for the LN levels included Dice similarity coefficient (DSC) and the
95th percentile Hausdorff distance in mm (HD95). For i-LNs the testing metrics
were DSC, sensitivity (SEN) and positive predictive value (PPV). SEN and PPV were
based on whether the predicted segmentations intersected with the ground truth
segmentations. Descriptive variables were reported as median with
inter-quartile range. The metrics of both networks were compared using the Wilcoxon
rank test.
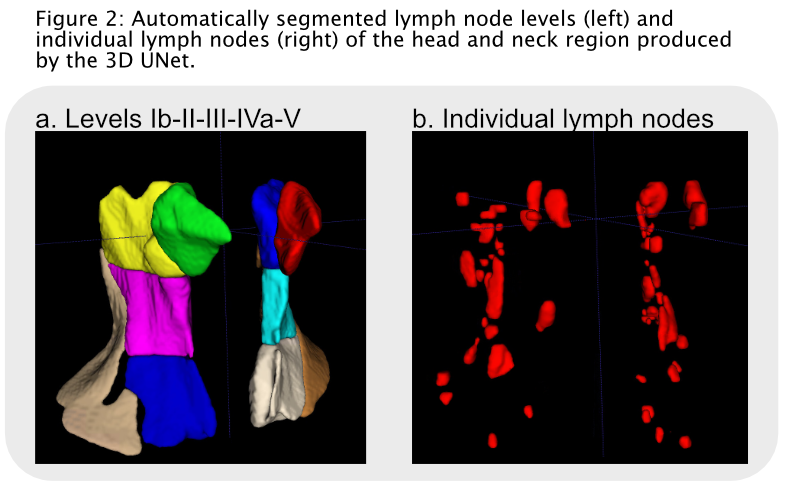
Results
The UNet
outperformed the DM network on both i-LNs and LN levels (Fig. 2). The UNet
produced higher DSC scores for segmentation of i-LNs compared to DM;
respectively 0.68 (0.60-0.72) versus 0.56 (0.53-0.68) (p=0.01). Comparable results were seen between both networks
regarding to SEN (UNet: 0.84 (0.75-0.88), DM: 0.89 (0.84-0.96), p=0.39).
The PPV was higher in favor of DM (UNet: 0.58 (0.56-0.61), DM: 0.66 (0.57-0.70), p=0.05).
For
most levels (II-V) on both sides of the neck the DSC and HD95 scores were significantly
better with the UNet. The median DSC and HD95 score for all LN levels were 0.73
(0.70-0.76) and
6.50 (5.80-7.30)
for UNet and 0.62 (0.59-0.65) and 8.29 (5.30-9.67) for DM. No difference was
found between both networks in the predicted segmentations of level Ia.
Conclusion
State
of the art 3D CNNs produce clinical acceptable automatic segmentations of i-LNs
and LN levels, bringing the irradiation of i-LNs closer to clinical
implementation. The UNet outperformed DM on both the segmentation of i-LNs and
LN levels with better matching contours. However, the overestimation of predicted
i-LNs was smaller while using DM compared to UNet. Still a high sensitivity is
the most important factor with respect to i-LNs irradiation, which were high
for both networks.