Evaluation of two commercial deep learning OAR segmentation models for prostate cancer treatment
PO-1776
Abstract
Evaluation of two commercial deep learning OAR segmentation models for prostate cancer treatment
Authors: Jenny Gorgisyan1, Ida Bengtsson1, Michael Lempart1,2, Minna Lerner1,2, Elinore Wieslander1, Sara Alkner3,4, Christian Jamtheim Gustafsson1,2
1Skåne University Hospital, Department of Hematology, Oncology and Radiation Physics, Lund, Sweden; 2Lund University, Department of Translational Sciences, Medical Radiation Physics, Malmö, Sweden; 3Lund University, Department of Clinical Sciences Lund, Oncology and Pathology, Lund, Sweden; 4Skåne University Hospital, Clinic of Oncology, Department of Hematology, Oncology and Radiation Physics, Lund, Sweden
Show Affiliations
Hide Affiliations
Purpose or Objective
To evaluate two commercial,
CE labeled deep
learning-based models for automatic organs at risk segmentation on planning CT
images for prostate cancer radiotherapy. Model evaluation was focused on
assessing both geometrical metrics and evaluating a potential time saving.
Material and Methods
The evaluated
models consisted of RayStation 10B Deep Learning Segmentation (RaySearch
Laboratories AB, Stockholm, Sweden) and MVision AI Segmentation Service (MVision,
Helsinki, Finland) and were applied to CT images for a dataset of 54 male
pelvis patients. The RaySearch model was re-trained with 44 clinic specific
patients (Skåne University Hospital, Lund, Sweden) for the femoral head
structures to adjust the model to our specific delineation guidelines. The
model was evaluated on 10 patients from the same clinic. Dice similarity
coefficient (DSC) and Hausdorff distance (95th percentile) was
computed for model evaluation, using an in-house developed Python script. The
average time for manual and AI model delineations was recorded.
Results
Average DSC scores
and Hausdorff distances for all patients and both models are presented in Figure
1 and Table 1, respectively. The femoral head segmentations in the re-trained
RaySearch model had increased overlap with our clinical data, with a DSC (mean±1
STD) for the right femoral head of 0.55±0.06 (n=53) increasing to 0.91±0.02 (n=10)
and mean Hausdorff (mm) decreasing from 55±7 (n=53) to 4±1 (n=10) (similar results
for the left femoral head). The deviation in femoral head compared to the
RaySearch and MVision original models occurred due to a difference in the
femoral head segmentation guideline in the clinic specific data, see Figure 2. Time
recording of manual delineation was 13 minutes compared to 0.5 minutes
(RaySearch) and 1.4 minutes (MVision) for the AI models, manual correction not
included.
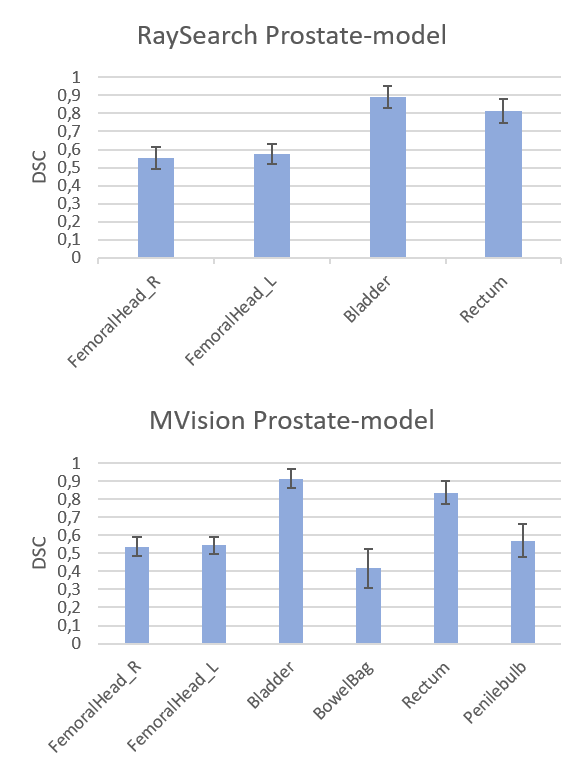
Figure 1. DSC
scores (mean values with 1 STD as error bars) for the RaySearch model (top) and
MVision model (bottom).
Table 1. Mean Hausdorff
distance ± 1 STD (mm) for different anatomical structures presented for both models.
| FemoralHead_R
n=53
| FemoralHead_L
n=53
| Bladder
n=54
| Rectum
n=53
| BowelBag
n=13
| Penilebulb
n=25
|
RaySearch
| 55±7
| 53±7
| 5±5
| 18±10
| - | - |
MVision
| 59±5
| 59±5
| 4±4
| 12±7
| 140±23
| 7±2
|
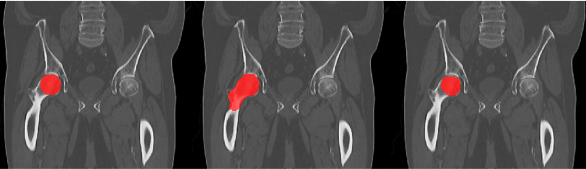
Figure 2. Femoral
head segmentation: clinical data (left), RaySearch original model result
(middle) and re-trained RaySearch model result (right). The clinical segmentation
includes only a sphere-like structure to represent the femoral head, whereas
the RaySearch segmentation in original model includes both femoral head and
neck.
Conclusion
Both AI models demonstrate good
segmentation performance for bladder and rectum. Clinic specific training data (or
data that complies to the clinic specific delineation guideline) might be necessary
to achieve segmentation results in accordance to the clinical specific standard
for some anatomical structures, such as the femoral heads in our case. The time
saving was around 90%, not including manual correction.