Authors: Robert Finnegan1,2,3, Vicky Chin4,5,6, Phillip Chlap4,3,2, Ali Haidar4,2,3, James Otton4,7, Jason Dowling8,1, David Thwaites1, Shalini Vinod4,2,6, Geoff Delaney6,4,5, Lois Holloway9,1,4,3
1School of Physics, University of Sydney, Institute of Medical Physics, Sydney, Australia; 2SWSLHD, Liverpool and Macarthur Cancer Therapy Centres, Liverpool, Australia; 3Ingham Institute for Applied Medical Research, Medical Physics, Liverpool, Australia; 4University of New South Wales, South Western Sydney Clinical School, Sydney, Australia; 5SWSLHD, Liverpool and Macarthur Cancer Therapy Centre, Liverpool, Australia; 6Ingham Institute for Applied Medical Research, Radiation Oncology, Liverpool, Australia; 7SWSLHD, Department of Cardiolog, Liverpool, Australia; 8CSIRO, Australian e-Health Research Centre, Herston, Australia; 9SWSLHD, Liverpool and Macarthur Cancer Therapy Centre, Sydney, Australia
Show Affiliations
Hide Affiliations
Purpose or Objective
Current risk models of radiation-related cardiotoxicity are based on whole heart doses. Cardiac substructure doses may have more predictive value, although further research is needed. Automatic segmentation enables analysis of large retrospective datasets, however, existing approaches do not provide a suitable solution for the highly variable imaging from multi-institute, multi-site clinical radiotherapy data. Additionally, delineation of smaller cardiac structures on CT scans remains a challenge for both manual and automatic methods. Our goal was to develop and implement open-source software to automatically delineate cardiac substructures accurately, consistently, and reliably.
Material and Methods
We designed a hybrid method using a deep learning (DL) model (nnU-Net) for whole heart delineation and a modified anatomically-guided multi-atlas technique for delineation of large cardiac substructures (chambers and great vessels), and geometry-based algorithms to delineate small structures (coronary arteries, cardiac valves, conduction nodes) [Fig. 1]. The DL network was trained using 300 CT scans and clinical heart contours from 150 breast cancer and 150 lung cancer patients. The heart and cardiac substructures were manually contoured and automatic segmentations were generated on an independent set of 30 CT scans (20 breast cancer and 10 lung cancer patients), including scans specifically selected for variations in imaging (e.g. artefacts, contrast CT). Segmentation accuracy was measured with Dice Similarity Coefficient (DSC), mean distance to agreement (MDA), and Hausdorff distance (HD), calculated between manual and automatic delineations. Values were compared to previously assessed inter-observer contouring variability (IOV). Four DL models (2D, 3D full-res., 3D low-res., ensemble) were evaluated based on accuracy and segmentation failure rate (DSC<0.8), with consideration for execution speed.
Fig. 1
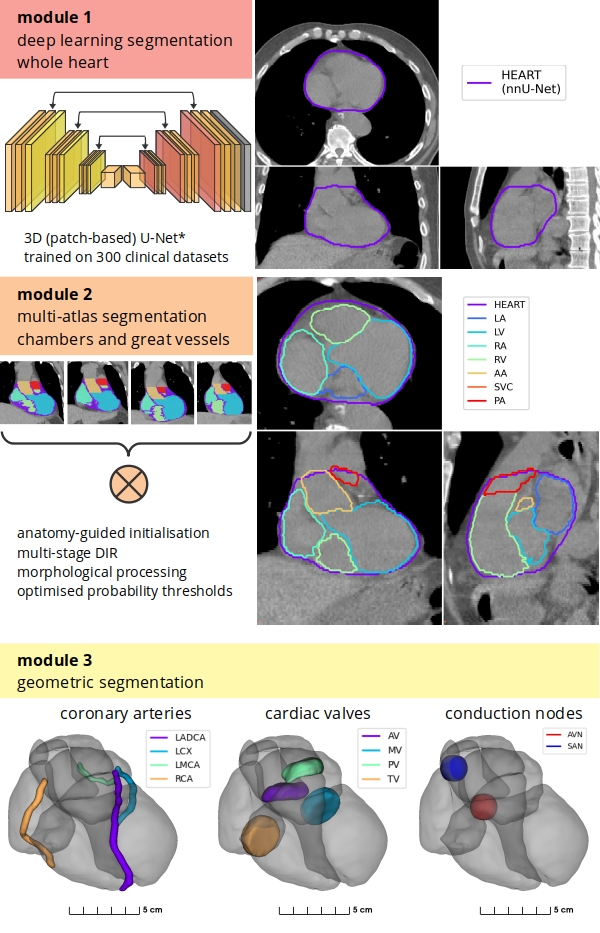
Results
All DL models produced accurate whole heart delineations (median DSC>0.931, MDA<2.06 mm, HD<12.6 mm); both 3D models provided the lowest failure rate (0/30 vs. 2/30 for 2D model) and the low-res. model had a faster execution speed. The ensemble model did not improve accuracy. Substructure segmentation accuracy was close to IOV [Fig. 2]. Geometric definitions for small structures improved consistency compared with our previous atlas-only method and provided anatomically realistic delineations of cardiac valves and coronary arteries.
Fig. 2
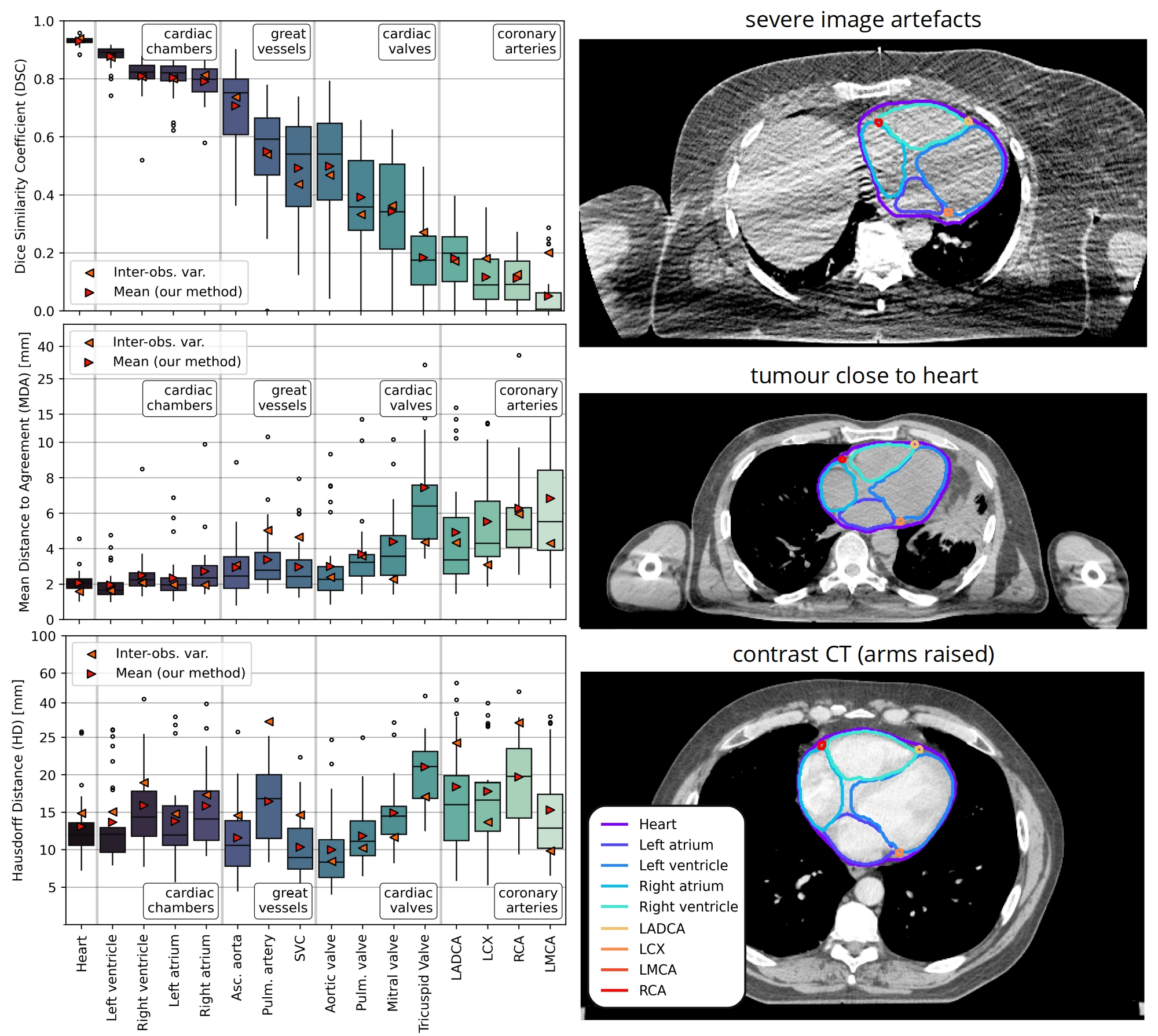
Conclusion
The hybrid method leverages the benefits of DL, atlas-based techniques, and geometric definitions to delineate cardiac substructures on CT imaging. Current work has validated the accuracy of dose metrics obtained using automatic segmentation on an independent dataset*. Our implementation provides a reliable and accessible model and will be used in future data-mining studies.
*V Chin et al (2021) Validation and clinical impact of a novel hybrid cardiac substructure automatic segmentation method, submitted to ESTRO 2022 (clinical track)