Privacy-preserving dashboard for clinical data using open-source federated learning infrastructure
Varsha Gouthamchand,
The Netherlands
PO-1062
Abstract
Privacy-preserving dashboard for clinical data using open-source federated learning infrastructure
Authors: Varsha Gouthamchand1, Gauri K2, Rajamenakshi Subramanian2, Ananya Choudhury1, Leonard Wee1, Andre Dekker1, Shwetabh Sinha3, Sarbani Ghosh Laskar3, Lohith Reddy4
1University of Maastricht, Faculty of Health, Medicine and Life Sciences, Maastricht, The Netherlands; 2Centre for Development of Advanced Computing (C-DAC), IT, Pune, India; 3Tata Memorial Hospital, Oncology, Mumbai, India; 4HealthCare Global Enterprises Limited, Oncology, Bangalore, India
Show Affiliations
Hide Affiliations
Purpose or Objective
Research on real-world cancer data
requires vast volumes of geographically distributed multi-institutional data to
be analyzed. It is not always feasible to centralize the data for statistical
analysis. “Distributed methods” are now making inroads into oncology research,
however confidentiality of individual human subjects remains the highest
priority. In keeping with Findable-Accessible-Interoperable-Reusable (FAIR)
data management principles, data schema and idiosyncratic coding should be
publishable as metadata, even though the data content itself is kept private.
Additional privacy is made possible by only exchanging statistical summaries,
but never any individual-level data. In this way, privacy is maintained without
relying exclusively on encryption or privacy-obscuring mechanisms (such as
differential privacy). This is known as privacy by design, since
re-identification of individual subjects from statistical summaries is a very
low risk, even where encryption and differential mechanisms are breached.
Material and Methods
We demonstrated a semi-automated
method for FAIR-ification of structured clinical data in four simulated geographically-separated
“data nodes”, each with one of four open-access HNSCC datasets. The datasets
were independent and had distinct schema with coding dictionaries. Individual
patient-level data were processed as Resource Descriptor Format (RDF). The anonymous
schema were separately extracted as an Ontology Web Language (OWL) resource. Each
OWL resource was shared, then individually mapped to a common oncology ontology
using an extra annotations layer above the raw data. In the same manner, three
private HNSCC datasets - from two Indian centers and one Dutch clinic - were processed
in the same manner. An interactive dashboard was created in Python to exploit
the Vantage6 distributed learning infrastructure. Distributed algorithms for
descriptive statistics of cohorts were transmitted through the Vantage6
infrastructure.
Results
The results from the algorithms
provided the summary statistics of the seven datasets, without combining any
individual patient-level data into a single repository (see Fig. 1). The
interactive display allows manual exploration of the data at deeper levels by
the researcher. The prototype visualization is interactive and can be easily
adapted to other use cases.
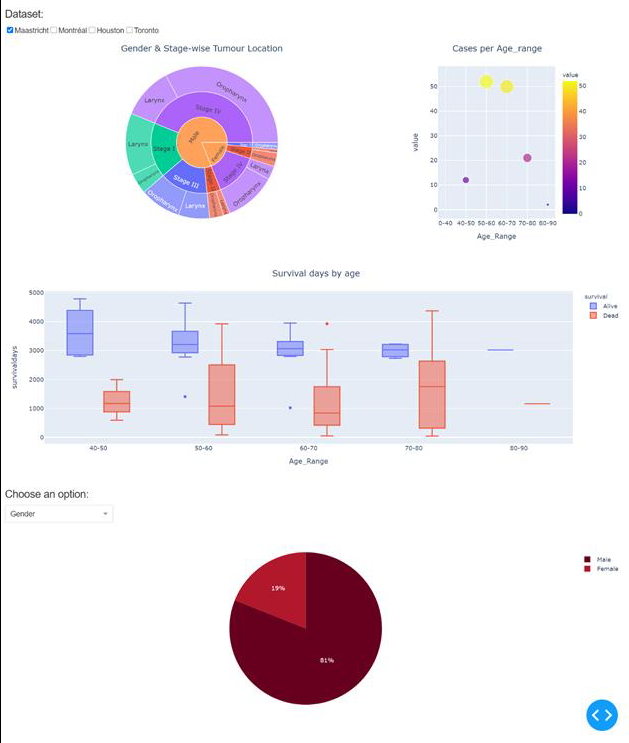
Fig 1. Privacy preserving distributed dashboard for
four public HNSCC datasets
Conclusion
An interactive distributed dashboard was
deployed through the open source federated privacy-preserving infrastructure, Vantage6.
The procedure was developed on four public datasets, then applied on three geographically
separated private datasets. Annotating datasets as FAIR data allows researchers
to explore and evaluate the case mix through a universal distributed dashboard
without breaching confidentiality of patient-level data. This allows institutions
to plan collaborations while retaining total control over their own data.