Style-based generative model to reconstruct head and neck 3D CTs
PO-1649
Abstract
Style-based generative model to reconstruct head and neck 3D CTs
Authors: Alexandre Cafaro1,2,3, Théophraste Henry3, Quentin Spinat1, Julie Colnot3, Amaury Leroy1,2,3, Pauline Maury3, Alexandre Munoz3, Guillaume Beldjoudi3, Léo Hardy1, Charlotte Robert3, Vincent Lepetit4, Nikos Paragios1, Vincent Grégoire3, Eric Deutsch3,2
1TheraPanacea, R&D Artificial Intelligence, Paris, France; 2Paris-Saclay University, Gustave Roussy, Inserm 1030, Molecular Radiotherapy and Therapeutic Innovation, Villejuif, France; 3Joint Collaboration Gustave Roussy - Centre Léon Bérard, Radiation Oncology, Villejuif-Lyon, France; 4Ecole des Ponts ParisTech, Research Artificial Intelligence, Marne-la-vallée, France
Show Affiliations
Hide Affiliations
Purpose or Objective
Generative Adversarial Networks (GAN), a deep learning method, has many potential applications in the medical field. Their capacity of realistic image synthesis combined with the control it enables, allows for data augmentation, image enhancement, image reconstruction, domain adaptation or even disease progression modeling. Compared to classical GANs, a StyleGAN modulates the output at different levels of resolution with a low-dimensional signature, allowing a control on coarse-to-fine anatomic structures when applied to medical data. In this study, we evaluated the potential of a 3D style-based GAN (StyleGAN) for generating and retrieving realistic head and neck 3D CTs, focused on a center zone prone to tumor presence.
Material and Methods
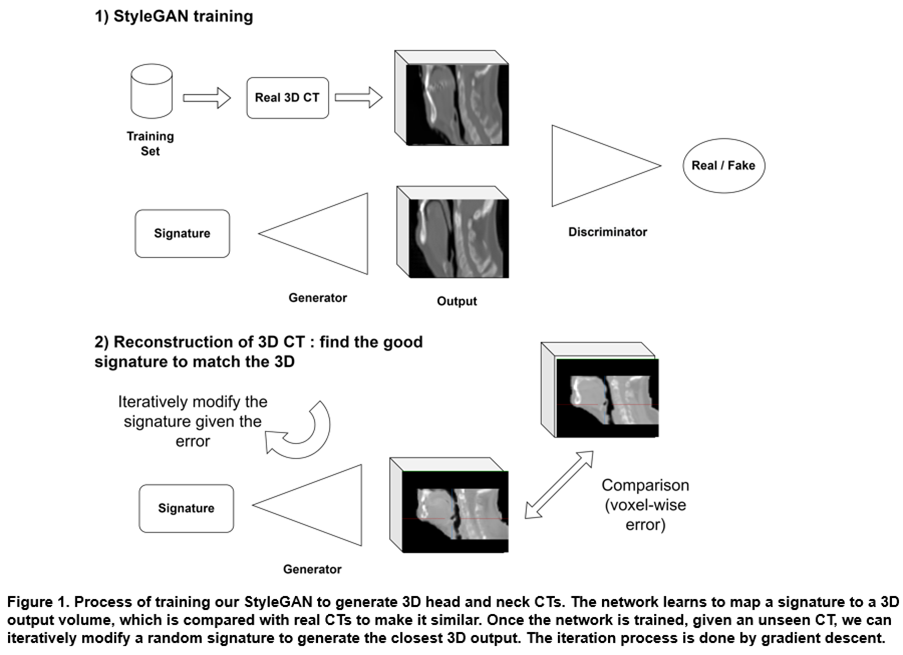
We trained our StyleGAN with a dataset constituted by 3500 CTs with head and neck cancers, from 6 publicly available cohorts from The Cancer Imaging Archive (TCIA) and private internal data. The dataset was splitted between 3000 cases for training and 500 for validation. CTs were focused on the head and neck center region around the mouth, with a small zone of 80x96x112 (1.3mm x 2.4mm x 1.9mm) due to high-memory requirements. After training, the model can generate synthetic but realistic 3D CTs from random "signatures". To evaluate the generative power of this model, we show that we can find signatures that generate synthetic 3D CTs very close to real ones. We present our process in Figure 1. We used 60 external patients undergoing VMAT treatment to benchmark the model with the mean reconstruction error, i.e., the normalized mean absolute error between the reconstructions and the real CTs.
Results
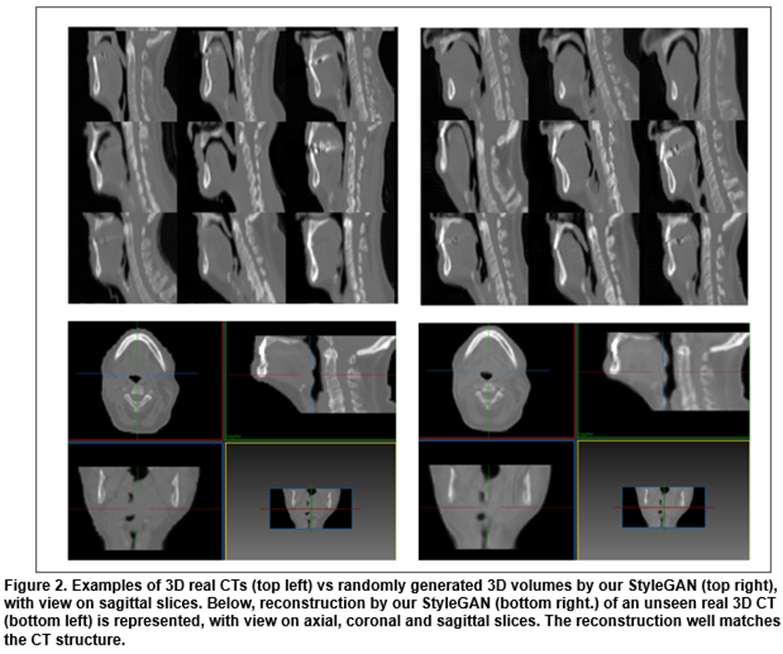
Training our StyleGAN took 2 weeks. After training, we randomly chose signatures which were given to the model to generate diverse and realistic 3D CTs as seen in Figure 2. We also show that given an unseen CT, we could generate the closest artificial version of it. This “reconstruction” is very close to the real case with almost the same level of details. Our model has a high capacity of retrieving the diversity of anatomies with fine details: on average, we achieve 1.7% (std. 0.5%) of reconstruction error on the 60 test patients. The signature that allowed us to generate the closest 3D output could be modulated afterwards to change the coarse-to-fine structures of the corresponding CT.
Conclusion
We have shown that our StyleGAN can learn to generate the spectrum of head-and-neck 3D CTs with a realistic and detailed anatomic structure. This enables a much finer and more diverse synthesis for data augmentation compared to using body phantoms. Our capacity to control the 3D generation at each resolution level allows us to well reconstruct real 3D CTs, enabling many medical image downstream tasks. In the future, we aim to increase the output resolution and . We will also explore whether the 3D CT synthesis process can be guided towards tumor generation in specific positions and sizes.