Robustness of nodal outlining software for breast radiotherapy
Alison McBride,
United Kingdom
PO-1644
Abstract
Robustness of nodal outlining software for breast radiotherapy
Authors: Alison McBride1, Rebecca Elt1, Colette Bennett1
1The Clatterbridge Cancer Centre, Physics, Liverpool, United Kingdom
Show Affiliations
Hide Affiliations
Purpose or Objective
Precise radiotherapy requires accurate outlining of clinical target volumes (CTV) and organs at risk (OAR). NICE guidelines recommend the treatment of the axillary, supraclavicular fossa (SCF), internal mammary nodal chain (IMN) and interpectoral nodes for node-positive invasive breast cancer. Currently our centre treats the axilla and SCF nodal volumes based on bony anatomy, IMN based on CT volumes and the interpectoral nodes are not treated. The protocol is being updated to adopt individual nodal level outlining based on CT volumes as recommended in ESTRO consensus guidelines.
There are commercial deep learning based autocontouring software (DLBAS) available that outline breast CTVs and OARs with the potential to reduce contouring time and inter- and intra-observer variability. The aim of this study was to assess the accuracy of the outlines produced by commercial DLBAS compared to gold standard clinician outlines.
Material and Methods
Sixteen patients previously treated for breast cancer were included in this study. For each dataset, clinicians manually outlined and peer reviewed a gold standard structure set in Eclipse. The primary and nodal level CTVs were outlined individually according to ESTRO guidelines. A whole nodal volume (LN) was created to encompass axillary levels 1-4 using the Boolean tool. OARs outlined included contralateral breast, the lungs, heart and brachial plexus.
Each dataset was sent to three commercial DLBAS – Mirada DLC Expert, Limbus AI and Siemens Organs. There were some differences between the availability of structures offered by each software. Mirada produced individual nodal outlines for axillary levels 1-4, interpectoral and IMN. Limbus AI generated LN and IMN structures. Siemens and Mirada created whole breast structures. All three produced OAR structures.
The resultant autocontours were compared to the gold standard using standard comparison metrics 3D Dice Similarity Coefficient (DSC), 95% Hausdorff Distance (95% HD) and added path length (APL).
Results
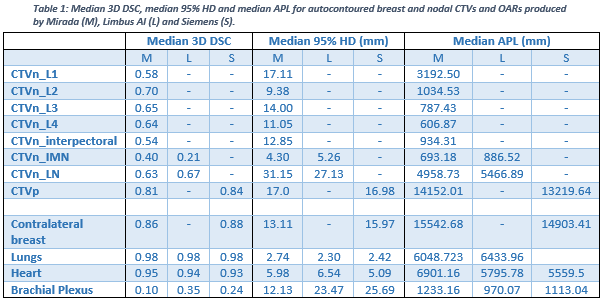
Table 1 contains the median geometric metrics for breast CTVs and OARs. For all structures the differences between systems were statistically insignificant apart from for the IMN metrics (p<0.004). The whole breast, lungs and heart autocontours were determined to have excellent agreement and could be used clinically. Nodal levels 1-4, interpectoral and LN volumes had moderate agreement and so require some editing, The IMN and brachial plexus had poor agreement so could not be used clinically without significant editing.
Conclusion
Autocontouring is possible for individual breast nodal volumes with moderate agreement to clinician outlines. Mirada is the only software investigated which produced individual structures and had better performance for small volumes such as the IMN. Whereas for larger structures such as the whole breast and the OARs tested there were comparable levels of performance between software. It is always important to review autocontours for suitability before approving for clinical.