Prediction of clonogenic survival following spatial fractionation - a novel 2D quadrat analysis
PO-2232
Abstract
Prediction of clonogenic survival following spatial fractionation - a novel 2D quadrat analysis
Authors: Delmon Arous1, Jacob Larsen Lie2, Nina Frederike Jeppesen Edin3, Eirik Malinen2
1Oslo University Hospital, Department of Medical Physics, Oslo, Norway; 2University of Oslo, Department of Physics, Oslo, Norway; 3University of Oslo , Department of Physics, Oslo, Norway
Show Affiliations
Hide Affiliations
Purpose or Objective
Spatially fractionated radiotherapy (SFRT) is an approach to deliver high local radiation doses in an ‘on-off’ pattern. How the clonogenic survival is affected by different SFRT patterns, in addition to local radiation dose, needs to be better characterized and appraised. The purpose of the current work was to analyze and predict clonogenic survival in vitro using a novel 2D quadrat Poisson regression analysis incorporating a generalized radiobiological model.
Material and Methods
A549 lung cancer cells cultured in vitro in T25 cm² flasks were irradiated using 220 kV X-rays with either an open, striped (5 mm openings and 10 mm blockings) or dotted (5 mm diameter openings and 18 mm center-to-center distance) fields. Nominal delivered doses (D) were 2, 5 and 10 Gy. Image segmentation was used to locate the centroid of surviving colonies (SCs) in scanned images of the cell flasks. Gafchromic™ film dosimetry (GFD) was employed to map the dose distribution in the flasks at each SC centroid. The paired, co-registered digital images with SCs and radiation dose were divided into 1 mm² quadrats. A modified linear-quadratic (LQ) model was used in a Poisson regression framework to analyze the SC count per quadrat. Here, D and D² where the principal explanatory variables with α and β as fit parameters. In addition, we introduced the nearest distance from the high-dose peak region to a given quadrat as an additional variable (peak distance, PD, η as fit parameter). The model was fitted to all SC data (open, striped, dotted) jointly. The Akaike information criterion (AIC) was used to evaluate model performance.
Results
We first compared results from conventional LQ analysis with Poisson regression on local quadrat SC data for open field irradiation. We obtained α=0.064±0.033 Gy‾¹ / β=0.011±0.007 Gy‾² and α=0.056±0.012 Gy‾¹ / β=0.014±0.002 Gy‾², respectively, indicating that the quadrat analysis gives the same results as conventional analysis but with lower errors. Furthermore, regressing over all survival data (open, striped, dotted), modified LQ-parameters gave α=0.033±0.008 Gy‾¹, β=0.025±0.001 Gy‾² and η=0.04±0.02 mm‾¹ (see Figure 1), with improved AIC.
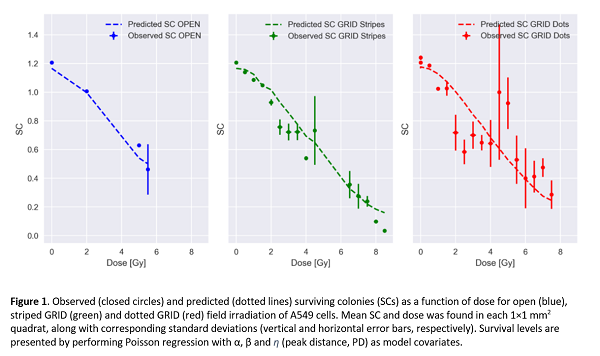
Conclusion
An advanced pipeline for mapping of SCs following GRID irradiation together with predicted survival levels was implemented. Dose is not the only valuable predictor for modulated fields, where PD was positively associated with survival for the given cell line.