Transcriptome profiling of tumour samples could predict radioresistance in glioblastoma patients
PO-2226
Abstract
Transcriptome profiling of tumour samples could predict radioresistance in glioblastoma patients
Authors: Sergi Serrano1, Gabriela Antelo1, Josep Balart2, Silvia Comas3, Cristina Ansón4, Carme Balañà5, Ana Maria Soto Cambres2, Izaskun Valduvieco1, Salvador Villà3, Meritxell Mollà1, Carles Gomà1
1Hospital Clínic de Barcelona, Radiation Oncology, Barcelona, Spain; 2Hospital de la Santa Creu i Sant Pau de Barcelona, Radiation Oncology, Barcelona, Spain; 3Institut Català d’Oncologia, Radiation Oncology, Badalona, Spain; 4Hospital de la Santa Creu i Sant Pau de Barcelona, Medical Physics and Radiation Protection, Barcelona, Spain; 5Institut Català d’Oncologia, Medical Oncology, Badalona, Spain
Show Affiliations
Hide Affiliations
Purpose or Objective
Glioblastoma (GB) is the most frequent and aggressive primary brain tumour in adults. Glioblastoma molecular subtypes, Classical (CL), Mesenchymal (MES) and Proneural (PN), have been correlated with prognosis. In the preclinical setting, CL and MES subtypes have been shown to be more radioresistant than the PN subtype. If confirmed in patients, the molecular subtype could predict the specific benefit of radiotherapy. The aim of this work is therefore to investigate whether GB molecular subtypes are associated with radiation resistance in vivo.
Material and Methods
We analysed the spatiotemporal pattern of recurrence (locoregional relapse and in-field relapse) in 55 GB patients treated with adjuvant radiotherapy (30 x 2 Gy, following EORTC contouring guidelines) using the Stupp’s regime included in a prospective multicentric study (GLIOCAT). We defined relapse following the RANO criteria and classified it spatially in 3 groups: in-field relapse (Dmin > 57 Gy), out-of-field relapse (Dmax < 30 Gy) and undefined (marginal relapse or no follow-up MR data). We used the Support Vector Machine (SVM) model for the transcriptomic classification of tumour samples. Patients with an undefined spatial pattern of recurrence were included in the progression-free survival (PFS) analysis but excluded from the in-field relapse analysis.
Results
The transcriptomic classification resulted in 26 CL, 12 MES and 17 PN. These groups were balanced with respect to the surgical procedure. Median PFS after radiotherapy was 4 months, with a median follow-up of 12 months. No differences in PFS were found between the different molecular subtypes (see figure 1a). However, we found differences in the in-field relapse-free survival between molecular subtypes, with a median time-to-event of 2.1 months (CL), 6.3 months (MES) and 6.9 months (PL) (figure 1b). These differences are statistically non-significant, probably due to reduced number of patients in each group (10 CL, 7 MES and 8 PL).
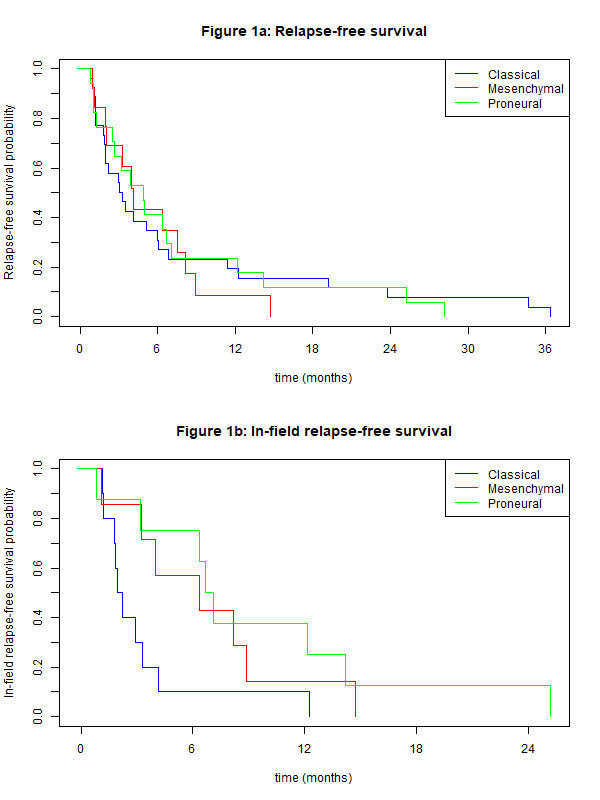
Figure 1: (a) Relapse-free survival probability and (b) in-field relapse-free survival probability, as a function of time (in months) for the different GB molecular subtypes: Classical, Mesenchymal and Proneural.
Conclusion
Although there is no difference in PFS among GB molecular subtypes, the results of this work seem to suggest that the Classical subtype relapses faster inside the radiotherapy field than the other subtypes. These findings would imply a higher radioresistance of the Classical subtype. Although these differences are statistically non-significant, they provide the rationale for further investigation in a larger patient cohort.