In silico model for low dose hyper-radiosensitivity linking mutation induction and cell survival
PO-2222
Abstract
In silico model for low dose hyper-radiosensitivity linking mutation induction and cell survival
Authors: Balázs Madas1, Balázs Madas2, Szabolcs Polgár3,4
1Centre for Energy Research, Environmental Physics Department, Budapest, Hungary; 2Budapest University of Technology and Economics, Department of Physical Chemistry and Materials Science, Budapest, Hungary; 3 Centre for Energy Research, Environmental Physics Department , Budapest, Hungary; 4ELTE Eötvös Loránd University, Doctoral School of Physics, Budapest, Hungary
Show Affiliations
Hide Affiliations
Purpose or Objective
Low dose hyper-radiosensitivity (HRS) and induced radioresistance (IRR) can be observed in the dose dependence of survival of many different cell lines.. While the phenomenon may have implications on both tumour control and normal tissue complication probabilities, its fundamental causes remain unclear. Earlier, it was hypothesised that low dose HRS is derived from a protective mechanism that has evolved to prevent genomic instability by removing those cells at risk of mutation. The objectives of the present study are to set up a computational model based on this hypothesis and to test its predictions with experimental data (1,2).
Material and Methods
The computational model considers mutagenic lesions occurring spontaneously during cell divisions (mean number is m in # per cell division) and mutagenic lesions induced directly by radiation (with a factor of μ in 1/Gy). Positions of cells are selected randomly on a virtual disk representing the culture dish. Cells emit signals to their environment with a strength proportional to the number of mutagenic lesions, which decreases with the distance. A cell goes into apoptosis if its mutagenic lesion number is higher than the sum of the mean number of mutations per cell division and the mean number of mutagenic lesions detected by the signal strength in the environment of the given cell. Surviving curves are determined supposing that the number of mutagenic lesions follows Poisson distribution with a mean proportional to absorbed dose. Monte-Carlo simulations were performed to fit the parameters (m and μ) to 101 experimental datasets.
Results
The computational model can reproduce the local minimum of surviving curves without introducing any arbitrary thresholds like a critical dose. The model can be fitted well to experimental datasets resulting in reasonable values for its two parameters. In general, the model fails to fit experimental data in those cases when the Induced Repair (IR) model, the most frequently used, descriptive mathematical model also fails.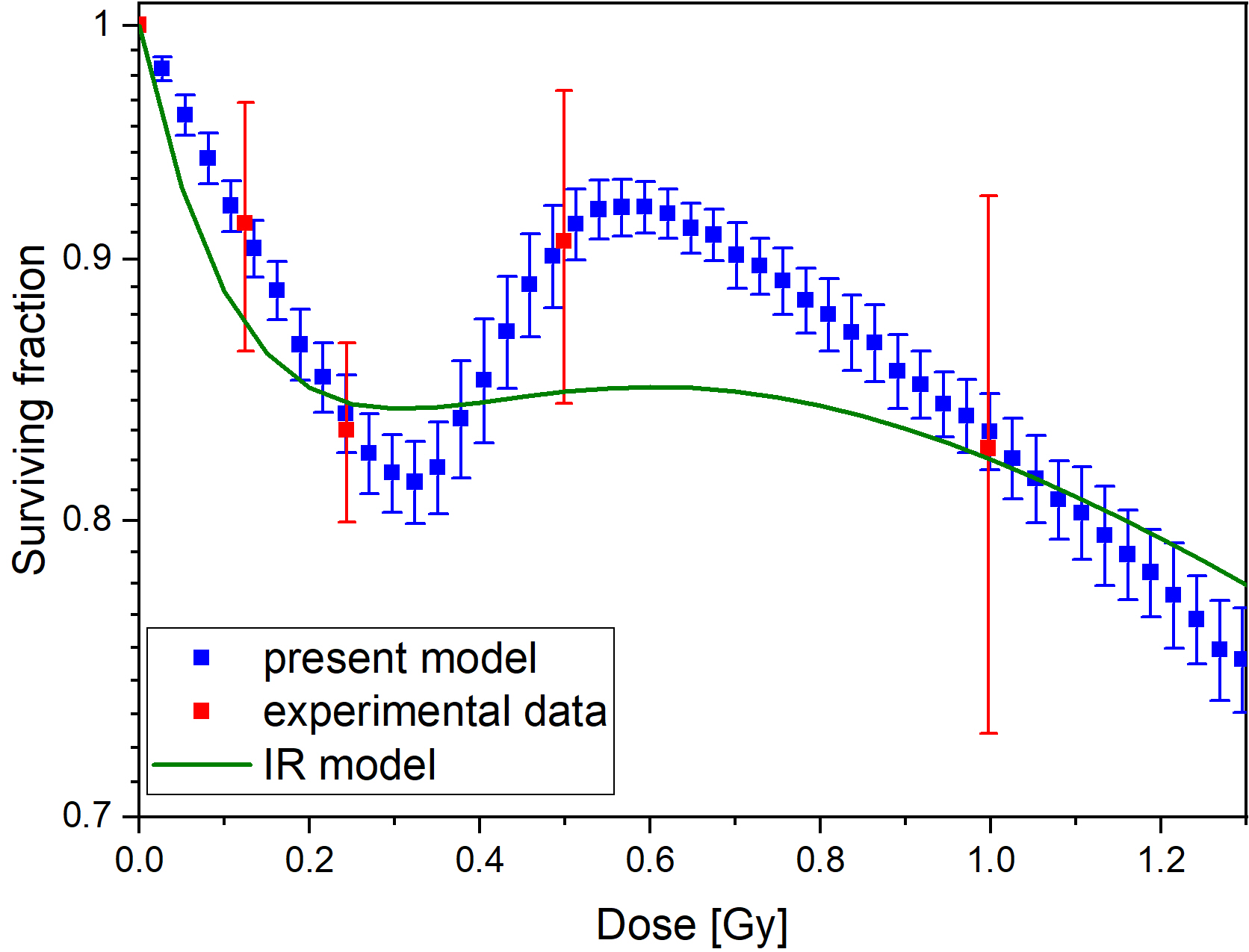
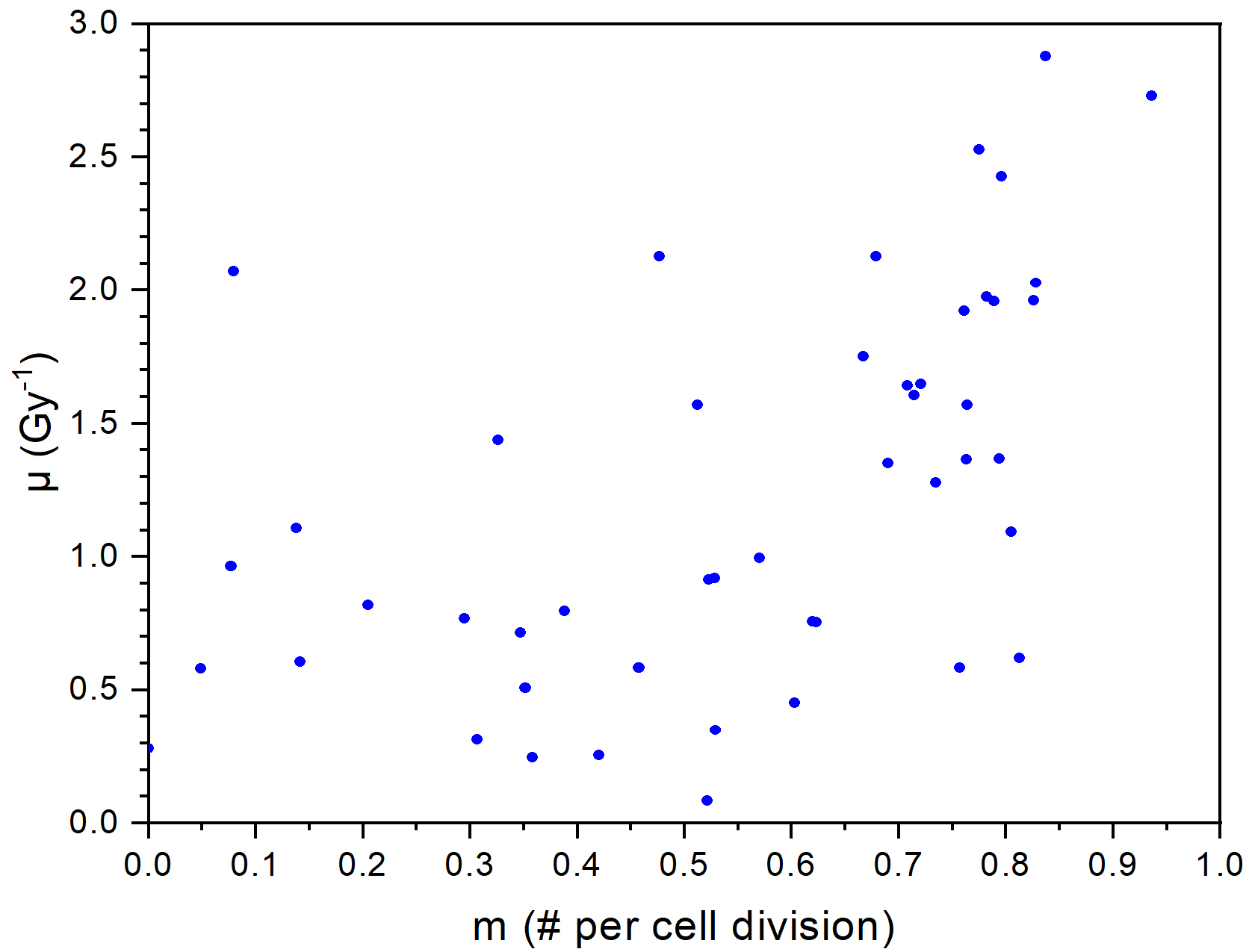
Figure 1. Comparison of fits of our model and the IR model for dataset 69 in ref. 2 (left panel), and distribution of parameter pairs obtained for experimental datasets of ref. 2 (right panel)
Conclusion
The computational model presented here provides a quantitative framework linking mutation induction and low dose hyper-radiosensitivity and induced radioresistance. In contrast with the IR model, the parameters of our model have clear biological meaning and can be measured independently on the colony forming assay. However, it provides similarly good fit to experimental data.
References
1. Polgár S, Schofield Paul N., Madas BG. 2022. Datasets of in vitro clonogenic assays showing low dose hyper-radiosensitivity and induced radioresistance. Sci Data 9(1):555. https://doi.org/10.1038/s41597-022-01653-3
2. Polgár S, Schofield Paul N, Madas BG. 2022. Data collection and analysis on low dose hyper-radiosensitivity and induced radio-resistance. https://doi.org/10.20348/STOREDB/1163