Pydicer: An open-source tool for conversion and analysis of radiotherapy imaging data
PO-1696
Abstract
Pydicer: An open-source tool for conversion and analysis of radiotherapy imaging data
Authors: Phillip Chlap1, Daniel Al Mouiee1, Shrikant Deshpande1, Robert Finnegan2, Janet Cui1, Vicky Chin1, Lois Holloway1
1UNSW, Ingham Institute & Liverpool and Macarthur Cancer Therapy Centres, Medical Physics, Liverpool, Australia; 2University of Sydney, Ingham Institute & Liverpool and Northern Sydney Cancer Centre, Medical Physics, Sydney, Australia
Show Affiliations
Hide Affiliations
Purpose or Objective
In many research projects that utilise radiotherapy imaging data (including structure sets and dose grids), a common first step is to prepare this data by converting it from DICOM into a suitable form for medical image analysis research. The NIfTI (Neuroimaging Informatics Technology Initiative) format is popular within the community and supported by many open-source Python libraries, including SimpleITK, PyRadiomics and nnUNet. We developed an open-source tool that solves many steps commonly undertaken within radiotherapy research projects to convert and analyse imaging data.
Material and Methods
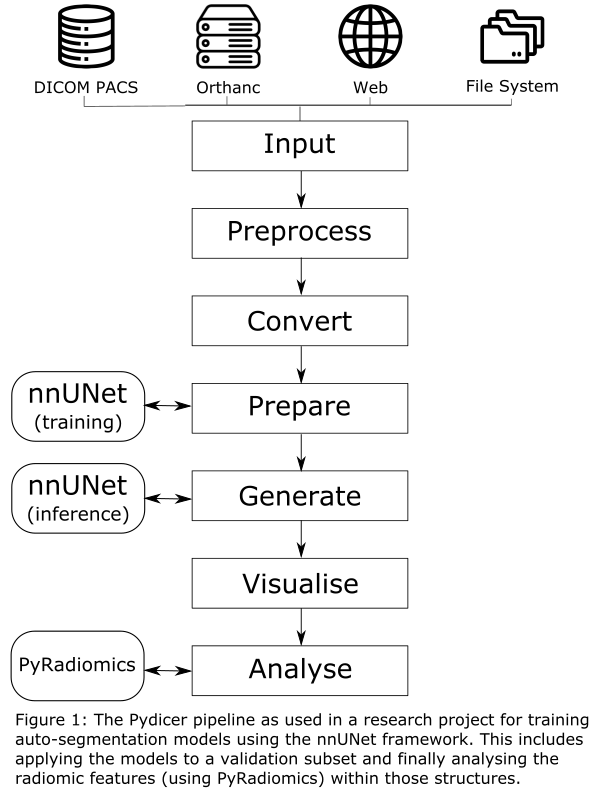
The Pydicer (PYthon Dicom Image ConvertER) tool (available open-source on GitHub: github.com/AustralianCancerDataNetwork/pydicer) is implemented using Python where several steps commonly undertaken in radiotherapy imaging research have been implemented that can be combined to produce a suitable pipeline for a given research project (figure 1).
A flexible input module provides functionality to fetch DICOM data from various common DICOM data sources. Additional modules deal with linking the DICOM objects (Preprocess), converting these to NIfTI format (Convert), cleaning up data into subsets (Prepare), producing additional data objects (Generate), visualising cross-sections (Visualise) and finally computing radiomic features or dose metrics (Analyse).
All converted data is stored within a well-defined directory structure with each object (image, structure set, dose, etc) saved to its own directory. This provides consistency with the DICOM standard to handle any dataset as well as flexibility to store accompanying data (such as radiomics features or dose volume histograms) alongside the NIfTI data. The tool tracks all converted data objects for each patient to streamline processing and analysis.
Results
The tool has been successfully utilised in several research projects including the analysis of two clinical trial datasets as well as cardiac sub-structure toxicities in a local dataset. Most notably an in-house project where auto-segmentation models are trained on local data using the nnUNet has been streamlined by utilising the Pydicer pipeline.
While some basic coding ability is required to use the tool, most logic has been abstracted into separate modules. This allows users who may have limited coding skills (such as medical physicists) to compose a suitable pipeline and process imaging data in bulk for their research. While difficult to quantify, this has shown a significant time saving over manual analysis using various commercial tools (e.g. MIM). The flexible nature of the tool enables the analysis of larger research datasets derived from our clinical data.
Conclusion
The Pydicer tool has demonstrated versatility across radiotherapy research projects that analyse imaging data. By being open source, the entire community can benefit from this tool and as features are added or bugs fixed these can be contributed to the tool for all to benefit.