Time-dependent diffusion MRI for non-invasive tumor characterization on a 1.5 T MR sim
Minea Jokivuolle,
Denmark
PO-2080
Abstract
Time-dependent diffusion MRI for non-invasive tumor characterization on a 1.5 T MR sim
Authors: Minea Jokivuolle1,2, Henrik Lundell3, Kristoffer Madsen3,4, Faisal Mahmood1,2
1Odense University Hospital, Laboratory of Radiation Physics, Department of Oncology, Odense, Denmark; 2University of Southern Denmark, Department of Clinical Research, Odense, Denmark; 3Danish Research Centre for Magnetic Resonance, Copenhagen University Hospital Amager and Hvidovre, Centre for Functional and Diagnostic Imaging and Research, Copenhagen, Denmark; 4Technical University of Denmark, Department of Applied Mathematics and Computer Science, Copenhagen, Denmark
Show Affiliations
Hide Affiliations
Purpose or Objective
Non-invasive methods for characterization of tumor microstructure are required to improve radiotherapy individualization. Time-dependent diffusion MRI (TDD-MRI) has shown potential to provide quantitative estimates of tissue microstructure, such as extracellular volume, membrane permeability, cellularity and cell size. However, so far TDD-MRI has mainly been investigated on high performance pre-clinical systems. Here, we investigated the capability of a clinical 1.5 T MR sim system to perform TDD-MRI to estimate cell sizes using asparagus as a model tissue, and demonstrated in vivo feasibility in human muscle tissue.
Material and Methods
Asparagus (asparagus officinalis) stems (N = 9) were cut to 4 cm pieces and stabilized in water. The left calf muscle of one healthy volunteer (M, 44 yrs, 81 kg) was imaged in vivo.
TDD-MRI was acquired on a 1.5 T MR sim (Ingenia, Philips) with a 45 mT/m gradient system, using spin-echo EPI readout (sequence parameters in Table 1). Measurements were repeated with increasing effective diffusion time (Td) to assess the time-dependency of the signal.
Table 1: MRI acquisition parameters.
| TR/TE (ms) | Matrix/resolution | b-values (s/mm²) | Gradient duration (ms) | Td (ms) | Scan time (mm:ss) |
| Asparagus | 3216/117 | 152x70x12/1x1x5 | 0, 500, 1000, 1500 | 24.0 | 32.0-62.0 | 01:27 |
| Muscle | 3216/95 | 64x61x10/2.5x2.5x5 | 0, 250, 500, 750 | 19.5 | 20.5-50.5 | 03:23 |
For validation, Monte Carlo simulations of MRI signal were performed for varying cell radii using intrinsic diffusivity of 2.1 µm²/ms for asparagus and 3.0 µm²/ms for muscle [1].
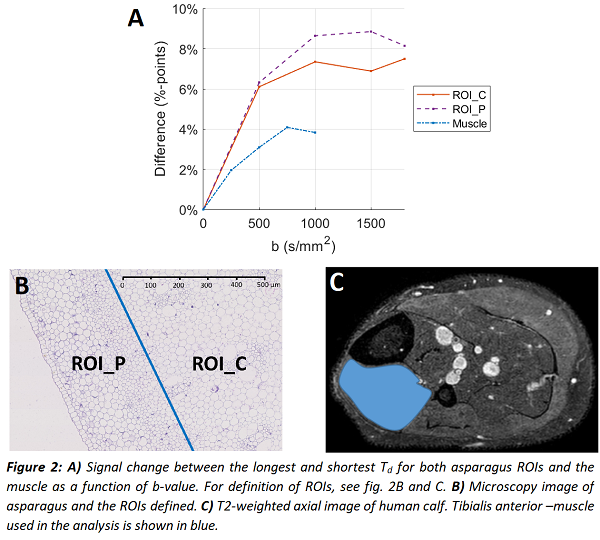
Analytical model for diffusion in cylinders [2] was fitted to the asparagus data to estimate cell radius (R) and diffusivity within two regions of interest (ROIs), corresponding to the central (ROI_C) and peripheral (ROI_P) area of asparagus stems.
Results
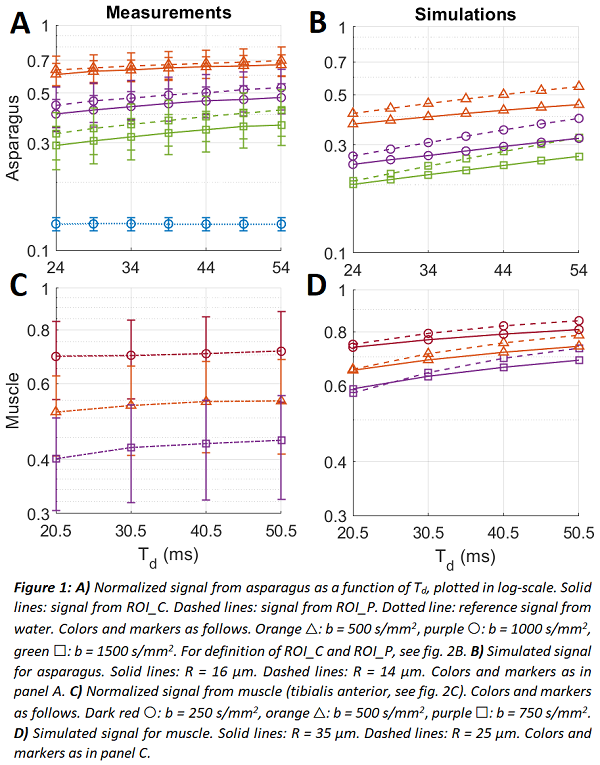
The measured diffusion signals from asparagus showed a clear time-dependency (fig. 1A), in agreement with the simulations (fig. 1B). The slope of the curves increased with b-value, indicating that the time-dependency is due to restricted diffusion within the cells. Signal changes up to 7.5 %-points in ROI_C and 8.9 %-points in ROI_P were observed (fig. 2A). The calf muscle showed a weaker, yet detectable effect (fig. 1C & 2A).
The analytical model estimated correctly a larger cell radius in ROI_C (16.38 µm) and a smaller one in ROI_P (14.75 µm) with estimated diffusivity of 1.40 µm²/ms in both ROIs. The difference to microscopy reference (18.52 µm) was small (~12%) in ROI_C, while for ROI_P difference to reference (7.69 µm) was large, likely due to partial volume effects (caused by large in-plane voxels).
Conclusion
We demonstrated the feasibility of TDD-MRI acquisition on a clinical 1.5 T MR sim with clinically relevant cell sizes. The results show promise for differentiation of tissues by cell size, which could help in characterizing tumor sub-regions for non-uniform radiation dose delivery.
[1] Hall et al., IEEE Trans Med Imaging, 28.9:1354, 2009
[2] van Gelderen et al., J. Magn. Reson., Series B 103.3:255, 1994