PhD Research Report
Novel concepts for automated segmentation to facilitate MRI-guided radiotherapy in head and neck cancer - PDF Version
Abstract
Treatment planning and response assessment of radiotherapy (RT) has seen a dramatic increase in the usage of magnetic resonance imaging (MRI). The information that is gained from the high soft-tissue contrast MR images can be used, for instance, to improve the segmentation of regions of interest (ROIs) on the CT scan for treatment planning. However, the current practice of manual segmentation is subjective and time-consuming, particularly for patients who have head and neck cancer (HNC), and therefore, the development of reliable auto-segmentation algorithms for organs-at-risk (OARs) and radiation targets is required. New methodologies based on machine learning offer ample opportunities to solve this problem.
My PhD project aimed to develop accurate and rapid auto-segmentation algorithms on MR images of HNC patients through the employment of established atlas-based algorithms and comparison of the results with deep learning-based methods. The work was divided into the design and implementation of auto-segmentation methods followed by extensive validation studies. For the latter, I developed a fully automated RT workflow that enabled validation on purely geometric features of the automatically generated contours and further analysis of the impact of geometric errors on key dosimetric features of a treatment plan.
A common challenge in medical image segmentation is the limited availability of data due to the associated cost of obtaining expert contours. Moreover, frequent updates of imaging protocols or scanners may prevent algorithms that are developed on existing databases from working well on newly-acquired images. Therefore, I designed domain adaptation methods that leveraged large databases from related application domains (i.e. annotated CT images) to tackle this problem (i.e. the contouring of MR images).
While both auto-segmentation strategies achieved clinically acceptable accuracies, atlas-based methods were slow and proved difficult to share between hospitals due to data-confidentiality issues. This is in contrast with deep-learning based models, where one can share a model which can be applied directly to new data without the need to share patient data. Additionally, deep learning methods can be used to alleviate the computational burden, as they generate contours within seconds. Moreover, in my study it was found that, when healthy tissue was infiltrated with irregular structures, deep learning was more accurate.
In conclusion, I could demonstrate that auto-segmentation was feasible and could change clinical practice. Moreover, I could show that domain adaptation strategies held promise in the mitigation of problems with small datasets in medical imaging and in the elimination of the need to acquire new annotated datasets for each change in imaging protocols.
What was the motivation for the topic of your PhD work?
The segmentation of OARs is essential for the treatment planning of RT. To minimise side effects, such as swallowing dysfunction or dry mouth, it is crucial to accurately localise these organs and take them into account during planning of the optimal configuration of beam intensities and arrangements.
Conventional RT treatments rely on the use of a snapshot of the patient’s anatomy for the full treatment course of about six weeks (for HNC patients). This process neglects potential changes in the patient’s anatomy due to weight loss, tumour or organ shrinkage, and swelling. With the introduction of MRI-guided treatment systems, visualisation of the soft-tissue contrast is improved, which is crucial for many structures in the head and neck.
Besides this visualisation advantage offered by MRI, and an extensive range of possible contrasts, it enables functional imaging which can be used to visualise surrogates, for instance for tumour metabolism, hypoxia, diffusion and perfusion.
To utilise fully the promises of these techniques, automation of the workflow is crucial as the current practice of manual segmentation is time-consuming and tedious. Besides, there is generally no ground truth and segmentation is hence subject to substantial inter- and intra-observer variabilities.
What were the main findings of your PhD?
There were three main findings in my PhD: 1) we can use automated segmentation methods for the purposes of treatment planning; 2) we need to take dosimetry into account during evaluation of contours; and 3) we can use generative adversarial networks (GANs) to generate substantially larger training sets than was previously possible. I will discuss these findings in more detail in the following paragraphs.
Following atlas-based and deep learning-based methods, I could demonstrate that it was feasible to segment OARs automatically with a geometric accuracy comparable to the measured inter-observer variability. However, atlas-based segmentation was not suitable for daily adaptations due to relatively long computation times. In addition to that, I found that it performed well for organs of similar shapes and locations for different patients, but could not cope when that was not the case.
The computational burden of conventional auto-segmentation algorithms could be alleviated with deep learning-based methods, with prediction times in the order of seconds or even sub-seconds. Furthermore, deep learning-based methods are more flexible and can be used to segment ROIs that vary in shape and location, such as involved lymph nodes or tumour volumes.
.png.aspx)
Segmentation examples: this figure provides four example cases for atlas- and
deep learning-based auto-segmented left parotid (LP) and right parotid (RP) glands (orange:
atlas-based, dark blue: deep learning-based, light blue: manual gold standard).
Each row shows a typical example, whereas the columns illustrate the axial,sagittal and coronal cross-sections, respectively.
Currently, auto-segmentation algorithms are commonly assessed by measures of volume overlaps or geometrical distances to a reference segmentation. However, for an application of auto-segmentation within an RT planning scenario, these geometric metrics are not necessarily meaningful. For example, a disagreement between manual and automated delineations in a region that undergoes a large dose gradient is likely to have a much more significant impact on an over-dosage to OARs or under-dosage to target volumes than in a region that only receives low doses. This emphasises the need for the establishment of additional, more meaningful evaluation metrics. I have demonstrated in my PhD that geometric measures alone are not sufficient to predict the impact of inaccurate segmentation on RT planning and that therefore the impact of segmentation errors on the planned dose distributions must be determined in order to evaluate thoroughly the auto-segmentation algorithms prior to their clinical implementation.
.png.aspx)
Overview of the fully automated validation workflow to evaluate auto-segmentation
algorithms in the context of RT. The highlighted part in orange shows the correlation analysis
to determine whether a geometric evaluation suffices as a surrogate for key dosimetric
features of a treatment plan. The top row illustrates an MR image, together with its gold
standard and auto-segmented ROIs. To perform a dosimetric analysis, we registered the
MR to its corresponding CT image via deformable image registration and used the resulting
deformable vector field to warp the segmented ROIs from the MR to the CT. The central part
of this figure shows the building blocks of the evaluation: the geometric and dosimetric evaluation, as well as the comparison with the inter-observer variability.
Furthermore, I was able to show that GANs hold tremendous promise in overcoming limitations due to the data sparsity that is generally faced in medical imaging. GANs can help in the exploration and discovery of the underlying structure of training data and can learn to generate new images. These approaches can be used to augment the small annotated datasets that are typically available in medical imaging.
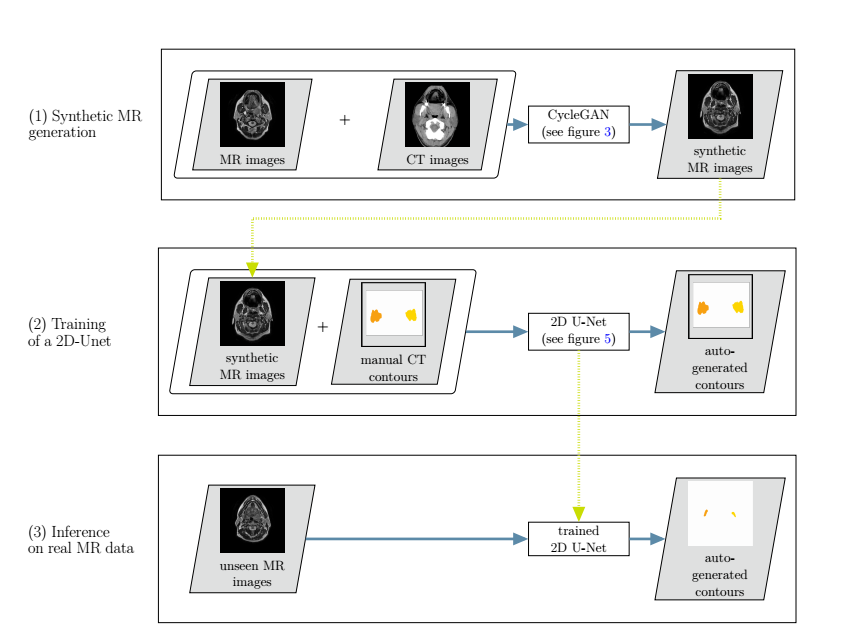
Overview of the developed cross-modality learning method: in the first step (top row),
synthetic MR images are generated through a generative adversarial network.The synthetic MR images are then fed into a 2D neural network, together with the annotations from the CT images (second row). In a third step, the trained network is applied to unseen real MR images (bottom row).
Can you comment on the impact of your work to the field?
I am convinced that adaptive RT will be one of the main driving forces for full automation of routine tasks in the clinical RT workflow in the near future, due to the necessity for full exploitation of a large quantity of imaging data. This will pave the way for automation, in particular of segmentation, in the general clinical RT workflow.
While deep learning-based algorithms are still “black boxes” to a large degree, it is easy for a human observer to verify the resultant automated delineation. For these reasons, auto-segmentation could be implemented in the clinic now, as a starting point for clinicians who then have the possibility to edit the proposed contours. I believe that auto-segmentation will gradually replace manual delineation, as clinicians gain confidence in the technique by observing its results. This would allow them to focus on different tasks.
Several vendors are already offering auto-segmentation algorithms in their commercial treatment planning systems with a recent trend towards deep learning-based methods. I believe that automated systems can reduce the number of human errors, improve consistency and create more time for other important tasks.
What was the most challenging part during your PhD?
A lack of delineated imaging data poses a problem for auto-segmentation methods for medical images. To perform well, most algorithms need a large amount of example imaging data. However, the receipt of annotated medical images is usually associated with a high cost, because the collection of large datasets of medical images is generally a time-consuming process. It involves the search for suitable data in large hospital systems with only moderately structured information and further processing (annotation) of the data by expert physicians. Patient consent is also required for use of the data. As a PhD student, I found it hard to obtain data from other institutes. I am grateful to the group of Professor Dave Fuller at the MD Anderson Cancer Center (Texas, USA) for helping me out with some of their datasets. I think it is essential that more hospitals share their data with each other to overcome this data problem.
In addition to a general lack of data, subsequent MR images are likely to change in appearance due to changes in image acquisition parameters or updates of MR scanners. Hence, auto-segmentation approaches that are developed on an existing image database may not work well on newly acquired images.
This lack of data was a recurrent theme in my PhD and, as I became aware that this was a common issue to varying degrees in medical imaging, I looked at solutions to this issue.
A second challenge was that initially, no one else in my department was working on deep learning-based methods, so I had to find expertise outside my department.
Will you stay in the field? What are your plans for the future?
Yes, I am still in the field of radiotherapy. I am now working on a very exciting project at the Imaging Lab of Varian in Switzerland, where I can apply some knowledge of maths, physics and radiotherapy, and am learning many more new skills, such as writing high-quality code.
Which institution were you affiliated to during your PhD?
I was working at the Institute of Cancer Research and the Royal Marsden Hospital in London, UK.
When did you defend your thesis and who was your supervisor?
My main supervisor was Professor Uwe Oelfke. I defended my thesis on the 5 September, 2019. It was a surprisingly pleasant experience as my examiners, Professor Coen Rasch and Professor David Hawkes, were both really interested in my work and we had very interesting discussions. At the end, I could not believe that four hours of discussion had gone by so quickly.
About the author
Jennifer Kieselmann received her Bachelor of Science degree in 2012 (thesis in the field of theoretical particle physics) and her Master of Science degree in 2014 (thesis on applying methods from differential geometry to dose calculation), both in physics at the Heidelberg University in Germany. After a nine-month internship at the Imaging Lab (ILab) of Varian Medical Systems in Baden (Switzerland), she did a four-year PhD in London at the Institute of Cancer Research, working on “Novel concepts for automated segmentation to facilitate MRI-guided radiotherapy in head and neck cancer”.
She is currently employed as a software development engineer at ILab. The results of her work have been or are currently in the progress of being published in high-impact journals and have been presented through oral communication at leading conferences.
.jpg.aspx)
Jennifer Kieselmann
Imaging Laboratory
Varian Medical Systems
Baden, Switzerland